Week 2- STAR Alignment
Arnab M -
Hi guys,
It’s Arnab and this is my second weekly update on my senior research project: Exploring the Genomic Effects of PNPLA7 Mutations on Cerebral Palsy through RNA Sequencing.
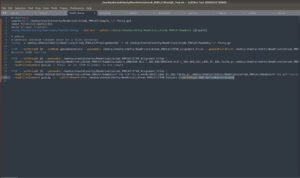
Last week my mentor and I designed a complete workflow for the early stages [FastQC→ MultiQC → STAR Alignment] that we’ve been successfully following. As the FastQC stage was completed we utilized another Bioinformatics tool called MultiQC. MultiQC summarizes all of the data collected in our numerous FastQC files and creates visual data graphs/plots (attached in the first image). These tools ensure our data is up to quality and ready to be put through our next stage: STAR Alignment. This is what I started working on in week 2 as STAR Alignment runs and processing can take up to a day, so it is crucial I did not mess up any code or organization of files for this step (attached in the second image).
STAR Alignment takes the Human Reference Genome (HG38) and compares it to all of our samples to align them together and annotate for differences. The coding to accomplish this was a step up from last week with far more intricacies and complications than last time, but in the end, I followed the STAR Alignment Manuals Coding instructions and successfully created a working code to run STAR which in turn produced BAM files, which I’ll discuss in next week’s blog.
As for more exciting news, the lab has granted me access to their wet lab to view in person some of their other ongoing neurological research on drosophila (fruit flies). I can’t wait to furthermore immerse myself in this translational research and move one step closer to giving children with neurological disabilities a better life.


Comments:
All viewpoints are welcome but profane, threatening, disrespectful, or harassing comments will not be tolerated and are subject to moderation up to, and including, full deletion.