The Reverse Transcriptase Quantitative Polymerase Chain Reaction
Hello everyone! This week, I will discuss the qPCR experimentation I completed to confirm the dot blot results from last week.
RT-qPCR is a Reverse Transcriptase quantitative Polymerase Chain Reaction. This method is designed to identify the amount of RNA encoded within a sample with one assay. The process consists of converting the RNA to complementary DNA (cDNA) and then amplifying the cDNA with specific primers.
Firstly, we extract the RNA and isolate it with the RNeasy Kit manufactured by QIAGEN. This involves cell lysis with Buffer RLT (RNA Lysis Buffer), RNA stabilization with 70% ethanol, binding the RNA to the silica membrane, and washing impurities with a wash buffer. To confirm the quality, we use a spectrometer. The spectrometer measures the concentration as well as the 260/280 ratio.
The 260 nm absorbance (A260) is the absorbance of nucleic acids at 260 nanometers, displaying the concentration of nucleic acids in the sample. The 280 nm absorbance (A280) is the absorbance of proteins at 280 nanometers, displaying the concentration of proteins in the sample. Thus, the 260/280 ratio illustrates the RNA concentration to proteins. Ratios between 1.9 and 2.1 for RNA indicate minimal protein contamination.
Secondly, we create the qPCR master mix for each sample. This contains the RNA template, primers, reverse transcriptase, DNA polymerase, Fluorescent Probes, Buffer System, and dNTPs (Deoxynucleotide Triphosphates). One set of the master mix is created without the RNA template, and one set is made with the primers for Beta Actin. The no template control tests the purity of the primers. Beta Actin is a housekeeping gene that tests the purity of the reagents. Another thing to note is that the template is always added last as it immediately starts reacting with the enzymes.
These mixes are added to the plate and put into the thermocycler. The thermocycler cycles between different temperatures. For RT-qPCR, we set each cycle to 50°C for 10 minutes (cDNA synthesis), 95°C for 2 minutes (enzyme activation), 95°C for 30-40 seconds (denaturation), 60-65°C for 20-35 seconds (Annealing), and 72-75°C for 5-10 minutes (Extension).
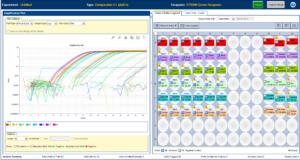
The qPCR machine measures the Cycle Threshold (CT). This is the cycle number at which the amplification of a gene crosses the predefined threshold value. Lower CT values suggest higher initial concentrations of the target gene. Generally, genes with a CT Value greater than 30 are considered minimal or nonexistent in the sample.
Ultimately, to confirm the results of the Dot Blot, I ran a qPCR (Real-time polymerase chain reaction), which evaluates the amount of mRNA coding for a specific protein. This experiment evaluated the mRNA coded for the HGF R receptor (c-MET) and Axl along with the housekeeping gene Beta Actin. This was performed on the Applied Biosystems® StepOne™ Real-Time PCR System. Although this does not confirm that all the mRNA is coded for the protein alone, with the Dot Blot, it emphasizes the results shown in Figure 5, in which Axl increased by ∼2-fold and c-MET increased by 8-fold.

Thank you for reading! I will discuss the growth curve next week and the properties of chemoresistant cell lines.
