Week 10: Gels, UV Plates, and Unexpected Bands
Daniel W -
After being out of town on Monday and Tuesday, I jumped right back into the lab midweek—and wow, things moved fast.
To quickly catch up new readers: for the past few weeks, I’ve been working on amplifying the promoter region of the cGAS gene, which plays a role in immune response. Because Primer-BLAST (a tool for finding specific primers) couldn’t find any primers specific enough to the region I wanted to amplify, I had to split the process into two rounds of PCR. Round 1 amplified a ~4787bp chunk of DNA, and Round 2 narrowed that down to the ~1919bp promoter region we actually need.
On Wednesday, I got to see the results of our Round 1 PCR on a gel electrophoresis—a method that uses electricity to separate DNA fragments by size through a gel. Although there were a few unexpected bands (which means extra DNA byproducts), we got a strong band right where we wanted: just below the 5000bp ladder.
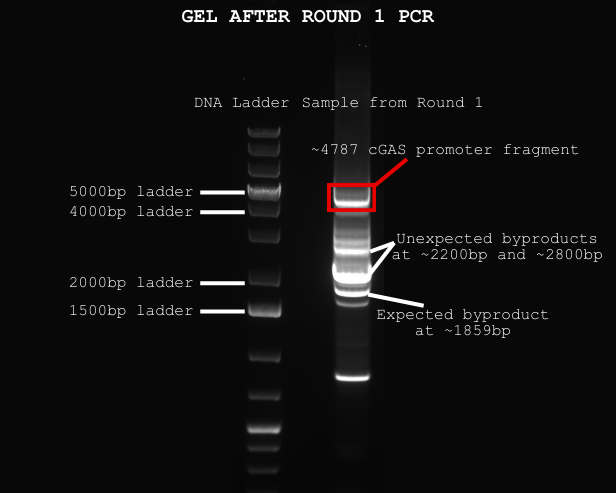
Since I wasn’t at the lab on Monday and Tuesday, Megan helped cut out the correct DNA band and clean it up with a PCI extraction, which uses chemicals like phenol and chloroform to remove unwanted proteins and enzymes from the DNA.
After analyzing our results, we ran Round 2 PCR using the cleaned Round 1 DNA as a template and a second set of primers. Since we were only copying a smaller region, we shortened the extension time. Afterward, we ran another gel to confirm if we got our ~1919bp cGAS promoter—and we did see a band at the right size! However, the band was quite large, and there were also two smaller, unexpected bands around 300–400bp. These might’ve been from non-specific primer binding, which was frustrating, but at least our main product was there.
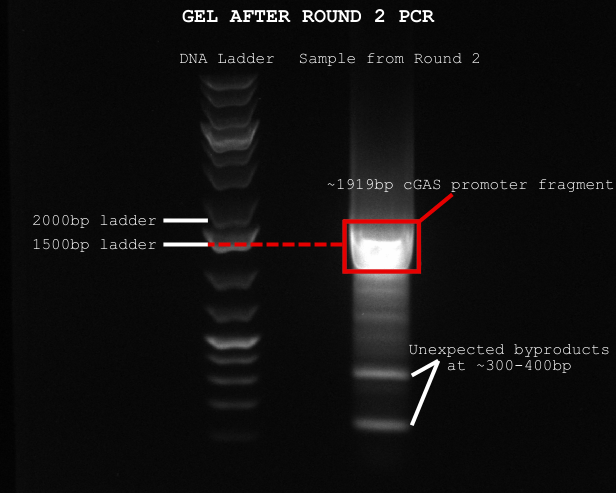
Under a UV plate, we carefully cut out the 1919bp band and started another PCI extraction and let it precipitate overnight.

On Thursday, we checked the concentration of our cleaned DNA using a Nanodrop, which uses UV light to measure DNA concentration and purity. The results showed a strong DNA yield, but unfortunately, the PCI cleanup hadn’t removed all the contaminants.

Since we wanted to be extra careful, we performed another round of PCI extraction to further purify the DNA and let it precipitate overnight.
On Friday, the Nanodrop results showed that the extra round of PCI extraction didn’t help much. We decided to move forward anyway and performed a restriction digest, which uses enzymes (in this case, NcoI and NheI) to cut the DNA at specific sequences. This will let us insert the promoter into our final plasmid—a circular piece of DNA we’ll use to test gene expression.
We also re-ran Round 1 PCR using a different DNA polymerase (the enzyme that builds new DNA during PCR) and tried several annealing temperatures in PCR to see if we could reduce the number of byproducts in future reactions. This enzyme is older but may bind more specifically to the DNA, which could help us get cleaner results next time.
Finally, I had a check-in with my senior project advisor about turning this into a poster presentation. We also talked about how I might include some of the physical products, like gels or diagrams, and even photos of myself working in the lab!
This week was full of progress and unexpected detours. I successfully completed both rounds of PCR for the cGAS promoter, verified the sizes via gel electrophoresis, and extracted the correct DNA using PCI. Although there were some byproducts and contaminants, I got a solid yield and began preparing the DNA for cloning into our luciferase plasmid. We also tested a new DNA polymerase to reduce byproducts in future runs. I’m getting close to the final cloning step—and even closer to turning this experience into a presentation I’m proud of!
– Daniel


Comments:
All viewpoints are welcome but profane, threatening, disrespectful, or harassing comments will not be tolerated and are subject to moderation up to, and including, full deletion.