Graphs Galore
Tisya o -
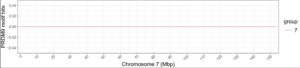
After much trial and error and help from my advisor, I successfully refined my PRDM9 graph for Haplotype 2 and created new graphs for Haplotype 1. As you can see below, there were very limited hits on the chromosome seven graph for Haplotype 2. I adjusted the x-axis scaling on the graph to better represent the data and better show the location of the hits. The concentration of hits in the first third of the chromosome seems to be consistent with the chromosomes’ five, eight, and nine’s PRDM9 motif hits positions. likely suggesting that most recombination and, therefore, chromosomal evolution occurs around the centromere. Unfortunately, there is not much data available about gelada centromeres since it is a very time-intensive and expensive process to create a reference genome. This means that I will not be able to analyze exactly how close these PRDM9 motif hits are to the centromere; instead, I am opting for rough estimates of the distance between the hits and the centromere using the data available to me.
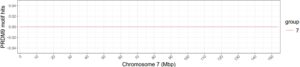

Interestingly, the Haplotype 1 chromosome seven graph showed zero hits. It is very probable that many PRDM9 hits in gelada are characterized by a different sequence of nucleotide base pairs that none of the discovered human motifs match, causing the program FIMO to skip over those sequences and resulting in no hits. On the other hand, this could be attributed to a lack of genetic recombination on this chromosome. Unless further study is done into mapping gelada-specific PRDM9 motif hits, our best option is to keep applying tests done on humans to geladas and hope that some pattern shows up.
Currently, I am working on creating a visualisation of the PRDM9 motif hits on all 21 chromosomes to include in my presentation while also working on completing my research paper.
